Cell culture
Human melanoma A375 cell line, murine melanoma B16F10 cell line, human colorectal cancer cell lines SW480, and human ovarian cancer cell lines SKOV3 were maintained in our laboratory. The cell lines were authenticated by STR profiling and regularly tested for mycoplasma contamination. Only mycoplasma-free cells were used for experiments. Cells were cultured in DMEM or RPMI-1640 medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (BI, Salt Lake City, UT, USA), and maintained at 37 °C in a humidified atmosphere of 5% CO2.
Stable NOS1 overexpression and nontargeted control cell lines were generated according to a previously reported method [57]. EX-I1936-Lv216–IRF7 (WT, C481A-MUT, Flag-tagged, human) and pLVX-mCherry-C1-IRF7 (WT, C435A-MUT, Flag-tagged, mouse) were designed and synthesized from IGEbio Technologies (Guangzhou, China).
The chemicals GSNO, N-PLA, L-NAME, and 1400 W were obtained from Cayman Chemical (Ann Arbor, MI, USA). Human and mouse IFNα (Sigma-Aldrich, St. Louis, MO, USA) was used to treat the cells for the indicated duration of time at a concentration of 1000 units per mL.
RNA extraction and quantitative PCR (qPCR)
Total RNA was extracted from cultured cells or tumor tissue, and cDNA was synthesized using RNAiso Plus reagent (Takara, Shiga, Japan) and PrimeScript RT kit (Takara), respectively. qPCR was performed on a LightCycler 96 System (Roche Life Science) using TB Green Premix Ex Taq II (Takara). All samples were normalized to the endogenous control GAPDH, and relative fold expression levels were calculated using the 2−ΔΔCt method. All experiments were performed independently at least three times, with all samples being analyzed in triplicate. The specific primers for RT-qPCR are as follows:
IFNα4 forward, 5’ -GACCTGCCTCACACTTAT-3’, IFNα4 reverse, 5’-AATCCTTCCTGTCCTTCA-3’, IFNβ1 forward, 5’ -GTTCCTGCTGTGCTTCTC-3’, IFNβ1 reverse, 5’-ATCTTCTCCGTCATCTCCAT-3’, IRF7 forward, 5’ -TGAGCGAAGAGAGCGAAGAG-3’, IRF7 reverse, 5’-CCAGTAGATCCAAGCTCCCG-3’, GAPDH forward, 5’ -CTACCCCCAATGTGTCCGTC-3’, GAPDH reverse, 5’-TGAAGTCGCAGGAGACAACC-3’, H2-K1 forward, 5’ -GGCAATGAGCAGAGTTTCCGAG-3’, H2-K1 reverse, 5’-CCACTTCACAGCCAGAGATCAC-3’, Cd74 forward, 5’ -CGAGAGCTGGATGAAGCAGT-3’, Cd74 reverse, 5’-GTTACCGTTCTCGTCGCACT-3’, H2-DMb1 forward, 5’ -GCTGTATCCATGGGCCGAAAAT-3’, H2-DMb1 reverse, 5’-AAGGTGTGTGGTTTGGGCTACT-3’, Tap2 forward, 5’ -CTGGCGGACATGGCTTTACTT-3’, Tap2 reverse, 5’-CTCCCACTTTTAGCAGTCCCC-3’, Tnf, forward, 5’-GGTGCCTATGTCTCAGCCTCTT-3’, Tnf, reverse, 5’-GCCATAGAACTGATGAGAGGGAG-3’, H2-M3 forward, 5’ -CTCCGCTACTACAACCAGAGTG-3’, H2-M3 reverse, 5’-TCTGAGCCATCATACGCAGCCT-3’, H2-T10, forward, 5’ -GGCTCGCAGACCCATGAAG-3’, H2-T10 reverse, 5’-GGCCTAGACAAGTTTTACAGGAC-3’, H2-T22 forward, 5’ -TCCCTTTGGGTTCACACTCG-3’, H2-T22 reverse, 5’-AGTCGTCCATGCTCTTGTTGT-3’, H2-D1, forward, 5’ -TGAGGAACCTGCTCGGCTACTA-3’, H2-D1 reverse, 5’-GGTCTTCGTTCAGGGCGATGTA-3’, Psme1 forward, 5’ -TCACTACCTGGTTGCAGCTACAG-3’, Psme1 reverse, 5’-GGTGTGAAGGTTGGTCATCAGC-3’, Tap1 forward, 5’ -GGACTTGCCTTGTTCCGAGAG-3’, Tap1 reverse, 5’-GCTGCCACATAACTGATAGCGA-3’, Psme2 forward, 5’ -TCCTCTGCACTTTCTTGCCACG-3’, Psme2 reverse, 5’-TGGGATAGGGATGTCCAGAGGA -3’.
Western blotting
Total protein was extracted using a Protein Extraction Kit (Fude, Hangzhou, China). Protein concentration in each sample was determined using the Bradford assay (Beyotime, Shanghai, China). Equal amounts of protein were separated on 8–15% SDS-PAGE gels and then transferred to PVDF membranes (Millipore, USA). The membranes were blocked with 5% BSA for 1 h. After blocking, the membranes were incubated with primary antibodies: anti-NOS1 (Abcam, Cambridge, MA, USA) at a 1:1000 dilution and anti-GAPDH (Proteintech, Wuhan, China) at a 1:5000 dilution. Following incubation with HRP-conjugated secondary antibodies, the immunoreactive bands were visualized using chemiluminescence (Bio-Rad, USA).
Coimmunoprecipitation (Co-IP)
Immunoprecipitates were obtained using the Co-Immunoprecipitation Kit (Cat. No.26149, Thermo Fisher), as described previously [57]. The assay was performed according to standard procedures.
S-nitrosylation detection assay
S-nitrosylated protein detection assays were performed as described previously [58]. Briefly, 100–250 μg protein lysates were extracted from A375 and B16 cells treated with IFNα (1000 U/mL), GSNO (100 μM), LNAME (1 mM), N-PLA (100 μM), or 1400 W(100 μM). Biotinylated proteins can be easily detected by biotin western blot or streptavidin precipitation, followed by western blotting.
CRISPR-Cas9-mediated genome editing, lentivirus production and cell line selection
IRF7-KO cells were obtained using the CRISPR-Cas9 system as described previously [Genome engineering using the CRISPR-Cas9 system]. The guide RNA (gRNA1: aaccatagaggcacccaag, gRNA2: acaagtgttggtaacaggt) was cloned into the Cas9 vector (NEWMOL, Synbio Technologies). Guide RNA-encoding plasmids were transfected into B16F10 cells for 48 h as described above.
Transfected cells were selected with G418 (300 ng/mL) to generate stable clonal lines from single cells, and individual clones were picked and cultured. Gene defects were identified by RT-PCR and immunoblotting. Lentiviral production was performed based on a previously described protocol [Production and purification of lentiviral vectors]. Stable IRF7-WT/MUT expression cell lines were generated by IRF7-KO cells infected with a lentivirus vector encoding IRF7-WT/MUT. Stable clones were selected with puromycin (1.5 μg/mL).
Tumor models
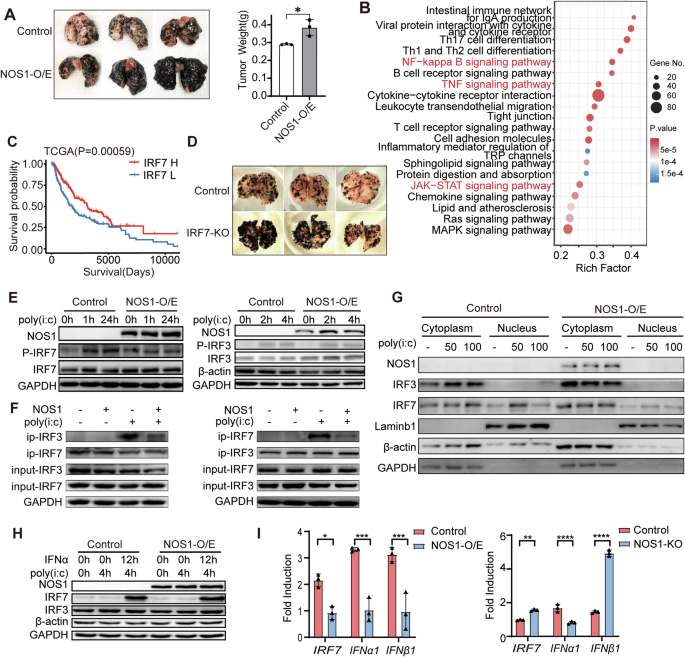
C57BL/6 mice and BALB/c-nu mice (Female, 4–5 weeks old) were all purchased from Guangdong Medical Laboratory Animal Center. To construct a lung metastasis model of melanoma, 5 × 105 B16F10 cells were intravenously injected into mice. After cell injection, the mice were randomly assigned to the experimental and control groups (5-8 mice per group), and they were then housed in SPF facilities on a 12 h light/ dark cycle until the end of the experiment. Mice were euthanized during days 15–17 postinjection, and lung tissue was isolated, photographed and then fixed with 4% formaldehyde for histological and morphometric measurements. In some cases, mice were sacrificed individually upon signs of metastatic distress and lung metastasis confirmed via histology and lung weight. The number of visible tumors in the lungs was counted separately, and fixed murine lungs were routinely processed and embedded in paraffin. Paraffin sections (5μm) were stained with H&E according to standard protocols, examined by microscopy and photographed.
Flow cytometry
For analysis of tumor-infiltrating lymphocytes, resected tumor tissues were cut into small pieces and then digested in collagenase Ⅱ (1 mg/ mL) and collagenase Ⅳ (2 mg/ mL) at 37 °C for 30 min. The mixture was filtered through a 70-μm strainer to prepare a single cell suspension. Cells were then washed twice with PBS and resuspended in PBS, and 1 × 106 cells were incubated with 3 μl antibody for 30 min at 4 °C in darkness. Wash the cells twice and perform the analysis on the FACS Calibur (BD Biosciences, USA). The anti-mouse CD3-PE-Cy7, CD8-FITC, CD45-APC-Cy7, F4/80-BV510, CD11b-BV650, IFN-γ-BV421, H2Kb-Alexa Fluor 647, CD279-PE, CD279- Granzymeb antibodies were all purchased from BD Biosciences. Data are represented as the percentage of lymphocytes as indicated.
Immunohistochemistry
Tumor tissues from B16-IRF7-WT/MUT tumor-bearing mice were collected in optimal cutting temperature (OCT) compound. Tumor tissues were sliced at a thickness of 4 μm. The tissue was fixed with 3.7% paraformaldehyde, and endogenous peroxidase was inactivated with 3% H2O2 at room temperature. Angiogenesis was detected with anti-CD4(1:1000 dilution, Abcam), anti-CD8(1:2000 dilution, Abcam) and anti-F4/80(1:100 dilution, Abcam) antibody. The level of angiogenesis was assessed at a magnification of ×200. Five randomly chosen fields per sample from each mouse were evaluated.
Gene expression profiling by RNA-seq
Total RNA was extracted using Trizol reagent (Thermo Fisher, 15596018) and purified using Dynabeads Oligo (dT) (Thermo Fisher) with two rounds of purification. A Poly(A) RNA sequencing library was prepared following Illumina’s TruSeq-stranded mRNA sample preparation protocol. RNA integrity was checked with Agilent Technologies 2100 Bioanalyzer. The mRNA was fragmented into short fragments using divalent cations under elevated temperature (94 °C) (NEB, cat. e6150). Then the cleaved RNA fragments were reverse-transcribed to cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, cat. 1896649). Paired-ended sequencing was performed on Illumina’s NovaSeq 6000 sequencing system. The reads containing sequencing adapters, sequencing primers, and sequences with q quality score lower than 20 were removed prior to assembly. The cleaned sequencing reads were aligned to the reference genome using the HISAT2 package, which built a database of potential splice junctions. Multiple alignments with a maximum of two mismatches were allowed for each read sequence. String Tie was used for assembling the aligned reads of individual samples. Transcriptomes from all samples were then merged to reconstruct a comprehensive transcriptome using a proprietary Perl script of LC Sciences (Houston, Texas, U.S.A.). FPKM reads and differential expressed genes were evaluated by StringTie and edgeR, respectively. The differentially expressed mRNAs and genes were selected with log2 (fold change) ≥0.26 or log2 (fold change) ≤−0.26, and with p values < 0.05.
Single-cell RNA sequencing (scRNA-seq) by Chromium 10x Genomics
Tumor tissues from B16-IRF7-WT/MUT tumor-bearing mice were cut into 2 mm pieces, and tumor tissues were prepared into a single-cell suspension with collagenase Ⅱ (1 mg/ mL) and collagenase Ⅳ (2 mg/ mL) at 37 °C for 30 min. Next, red blood cells were removed with red blood cell lysis buffer, and the single-cell suspensions were resuspended in flow staining buffer. Finally, the cells were treated with an Fc blocker to reduce the nonspecific binding of the Fc receptor and antibodies. The cells were labeled with rat anti-mouse BV510CD45 and 7-AAD (Cat. No: 51-65875X; BD Pharmingen). A total of 5 × 105 live immune cells (CD45 positive and 7-AAD negative) were sorted and resuspended in sample buffer for single-cell capture. Live CD45+ immune cells were captured using a BD RhapsodyTM system and lysed.
The overall cell viability was confirmed to be above 90%. Single cell suspensions were counted using a hemocytometer/Countess II Automated Cell Counter and concentration adjusted to 700–1200 cells/mL. Single-cell suspensions were loaded to 10x Chromium to capture 8000 single cells according to the manufacturer’s instructions of 10x Genomics Chromium Single-Cell kit (V3). The following cDNA amplification and library construction steps were performed according to the standard protocol. Libraries were sequenced on an Illumina NovaSeq 6000 sequencing system (paired-end multiplexing run, 150 bp) by LC-Bio Technology (Hangzhou, China) at a minimum depth of 20,000 reads per cell.
Bioinformatics and computational biology analyses
Sequencing data were de-multiplexed and converted to FASTQ format using Illumina bcl2fastq software (2.20). Sample demultiplexing, barcode processing and single-cell gene counting were initially processed using CellRanger (7.1) and scRNA-seq data were aligned to Ensemble genome GRCm38 reference genome, a total of 20529 single cell captured from 3 B16-IRF7WT mice and 3 B16-IRF7MUT mice were processed using 10X Genomics Chromium Single Cell Solution. The Cell Ranger output was loaded into Seurat (3.1.1) for dimensional reduction, clustering, and analysis of scRNA-seq data. Overall, 18,373 cells passed the quality control threshold: all genes expressed in less than 1 cells were removed, number of genes expressed per cell >500 as low and
Bioinformatic analysis was performed using the OmicStudio tools at https://www.omicstudio.cn/tool. DEGs were screened by R statistical package software EdgeR (Empirical Analysis of Digital Gene Expression in R, http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html) with Fold Change >1.2 or < −1.2, FDR < 0.05. Kyoto Encyclopaedia of Genes and Genomes pathway enrichment analyses of DEGs were performed by the KOBAS online database (http://kobas.cbi.pku.edu.cn/kobas3).
Additionally, gene set enrichment analysis (GSEA) was performed using the “fgsea” package in R. We selected two GSEA databases for our analysis: hallmark gene sets and KEGG gene sets. Moreover, MetaCore software was used for pathway analysis of significant genes. The cluster annotations were established based on signature genes that were approved by previous studies. Highly expressed genes in CD8 + T cells were assessed using f tests, t tests, and p values < 0.05. A list of differentially expressed genes was imported into the Metacore software to construct signaling pathways. The significance cut-off point for enrichment of a pathway was a p value < 0.05.
Statistical analysis
GraphPad Prism Software (version 9.0) was used for statistical analysis and preparation of most figures. Flow cytometry data analysis was carried out using FlowJo (version 10.0). ImageJ (version 2.3.0) was used for quantitative comparison of positive signals in Immunohistochemistry staining images. The choice of tests was based on the distribution and variance of the data. Normality was assessed using the Shapiro–Wilk test, and homogeneity of variances was tested using Levene’s test. For normally distributed data with equal variances, comparisons between two groups were made using the unpaired two-tailed Student’s t test. For non-normally distributed data, non-parametric tests (e.g., Mann–Whitney U test) were used. Two-way ANOVA with Tukey’s multiple comparison test was performed for multiple group comparison. The survival curves were performed by the Kaplan–Meier method and compared by the log-rank (Mantel–Cox) test, as indicated in the figure legends. The results are presented as mean ± SD unless otherwise noted. p < 0.05 was considered statistically significant. Mice were assigned to groups randomly. Analyses were not blinded, and no datapoints were removed from the analyses.