Mice
Male and female C57BL/6J (B6, strain 000664), B6.SJL-Ptprca Pepcb/BoyJ (B6 CD45.1, strain 002014) and B6(Cg)-Tcf7tm1Hhx/J (Tcf7-GFP reporter, strain 030909) mice were purchased from The Jackson Laboratory. Bach2loxP/loxP and Bach2loxP/loxP CreERT2 mice were described in our previous study15. All mice used in the experiments of this study were between 6 and 12 weeks old, age and sex matched and maintained on a B6 genetic background. Mice were fed an irradiated 2916 Teklad Global Rodent Diet and were kept in specific pathogen-free facilities with a 12-h light/12-h dark cycle. The room temperature range was 20–25 °C. Humidity was maintained at 30–70%. All animal procedures were approved by the Institutional Animal Care and Use Committee at University of Texas Southwestern Medical Center under protocol numbers 103162 and 103111.
Tumor inoculation
Syngeneic tumor cell lines E2A–PBX1 B cell ALL, B16-CD19 melanoma and 9464D-GD2 neuroblastoma are in a B6 background and have been described in previously published studies46,58,60. E2A–PBX1 cells were intravenously inoculated at 1 × 106 per mouse, whereas B16-CD19 and 9464D-GD2 cells were subcutaneously injected at 1 × 106 or 2 × 106 per mouse, respectively. Mice were euthanized if the tumor size exceeded 2 cm in diameter, if the tumor burden exceeded 10% of body weight or if animals exhibited >20% loss in body weight. Mice were also euthanized if tumors caused notable clinical signs, including pain, impaired mobility or evidence of metastasis. At no point during the study were these humane endpoints exceeded.
Transfection and retroviral transduction
For transducing mouse T cells, retroviral plasmid, pCL-Eco plasmid, Opti-MEM (Thermo Fisher Scientific) and TransIT-293 Transfection Reagent (Mirus Bio) were mixed, incubated and added to HEK293T cell cultures (ATCC, CRL-3216). Two days after transfection, culture supernatants with retroviruses were collected and centrifuged to remove cell debris. Mouse T cells activated by plate-bound anti-CD3 and anti-CD28 were transduced by a mixture of retroviruses and 8 µg ml−1 polybrene via spinoculation at 32 °C for 90 min. Anti-mouse CD19 CAR, described in our previous study16, was derived from the monoclonal antibody 1D3 and contains sequences from mouse CD28 and CD3ζ. An MSCV retroviral plasmid expressed anti-mouse CD19 CAR and a truncated huEGFR separated by a P2A peptide. The MSGV plasmid with an anti-GD2 CAR insert was a generous gift from C. Mackall (Stanford University) and was used in our previous study38. pMIG with a human BACH2 OE cassette was described by us previously15. A previously described BACH2–ERT2 cassette61 was inserted into pMIG to generate a retroviral construct with tamoxifen-responsive BACH2. To generate DD–BACH2 pMIG plasmid, a destabilization domain derived from a mutant FKBP12 (ref. 57) was fused to BACH2. A gRNA targeting Junb (ACGCAGCGGCGGGATACGGT) was cloned into SL21-VEX plasmid, a gift from J. Shi (University of Pennsylvania). For transducing human T cells, retroviral supernatants were generated using the 293GP packaging cell line (Takara, 631512). Briefly, 293GP cells were co-transfected with retroviral and RD114 envelope plasmids using TransIT-293 Transfection Reagent (Mirus Bio) in Opti-MEM (Thermo Fisher Scientific). Viral supernatants were collected 48 h after transfection and centrifuged to remove cellular debris. Human T cells purified from the peripheral blood mononuclear cells of healthy donors with an EasySep Human T Cell Isolation kit (StemCell Technologies) were activated with Dynabeads Human T-Expander CD3/CD28 (Gibco) at a 3:1 bead:cell ratio and transduced on days 2 and 3 after activation. Non-tissue culture-treated 24-well plates were coated overnight at 4 °C with RetroNectin (25 μg ml−1; Takara) in PBS, washed twice with PBS and blocked with 2% bovine serum albumin for 15 min. Retroviral supernatants were added, and plates were centrifuged at 2,000g for 2 h at 32 °C before the addition of T cells. On day 4 after activation, beads were removed by magnetic separation using an EasySep Magnet (StemCell Technologies).
Radiation and cell transfer
One day before CAR T cell transfer, mice were sublethally irradiated (500 cGy) as previously described16,38,47. CD19 CAR T cells or GD2 CAR T cells were adoptively transferred into mice through tail vein injection.
Tamoxifen treatment
As we previously described15, Bach2loxP/loxP CreERT2 mice were intraperitoneally injected with tamoxifen dissolved in sunflower seed oil to induce deletion of the floxed genomic segment. Control (Bach2loxP/loxP) mice were also treated with tamoxifen.
Flow cytometry and cell sorting
Flow cytometry and fluorescence-activated cell sorting (FACS) in this study were performed with the following antibodies and dyes. Anti-mouse CD45.1–Alexa Fluor 700 (A20, 1:200), anti-mouse CD4–FITC (RM4-5, 1:200), anti-mouse CD8a–BV605 (53-6.7, 1:200), anti-human EGFR–PE/Cy7 (AY13, 1:200), anti-mouse PD-1–PE (RMP1-30, 1:200), anti-mouse KLRG1–BV421 (2F1, 1:200), anti-mouse TIM3–APC (RMT3-23, 1:100), anti-mouse TIM3–BV605 (RMT3-23, 1:100), anti-mouse TIM3–PE/Cy7 (RMT3-23, 1:100), anti-mouse CX3CR1–FITC (SA011F11, 1:200), anti-mouse CX3CR1–PE/Cy7 (SA011F11, 1:800), anti-human CD4–PerCP/Cy5.5 (OKT4, 1:400), anti-human CD366 (TIM3)–PE (F38-2E2, 1:200), anti-human CD197 (CCR7)–PE/Cy7 (G043H7, 1:200), anti-human CD8–Alexa Fluor 700 (SK1, 1:400), anti-human CD39–APC/Cy7 (A1, 1:200), anti-human CD127 (IL-7Rα)–APC/Cy7 (A019D5, 1:200), anti-human CD62L–BV421 (DREG-56, 1:200) and anti-human CD45RA–BV785 (HI100, 1:200) were purchased from BioLegend. Anti-mouse CD62L–PerCP/Cy5.5 (MEL-14, 1:200), anti-TOX–PE (TXRX10, 1:100), goat anti-rabbit IgG (H + L) APC (1:500), anti-human CD223 (LAG3)–PE (3DS223H, 1:200), anti-human CD279 (PD-1)–PE/Cy7 (eBioJ105 (J105), 1:200), LIVE/DEAD Fixable Aqua Dead Cell Staining kit (1:250) and LIVE/DEAD Fixable Near-IR Dead Cell Stain kit (1:400) were purchased from Thermo Fisher. Anti-mouse granzyme B–BV421 (GB11, 1:200) and anti-active caspase-3–FITC (C92-605, 1:10) were from BD Biosciences. Anti-TCF1 (C63D9, 1:200) was purchased from Cell Signaling Technology. Flow cytometry was performed with a Cytek Aurora using SpectroFlo (v3.0.3). FACS was performed with a BD FACSAria II using BD FACSDIVA (v9.0.1). Analysis was performed with FlowJo 10.10.0.
Western blotting
Proteins were extracted from cells using RIPA Lysis and Extraction Buffer (Thermo Scientific) supplemented with Protease Inhibitor Cocktail (Thermo Scientific) and used for SDS–PAGE. The proteins were subsequently transferred to a PVDF membrane (Fisher Scientific) using a wet transfer method. The blots were blocked with 5% bovine serum albumin in Phosphate-Buffered Saline with Tween 20 and probed overnight at 4 °C with primary antibodies to BACH2 (1:2,000; Abcam, 7A4) and β-actin (1:1,000; Cell Signaling Technology, 8H10D10). Following washing, the blots were incubated with secondary horseradish peroxidase-conjugated antibodies (Cell Signaling Technology) at a 1:5,000 dilution for 60 min. Protein bands were visualized using enhanced chemiluminescent substrate (Thermo Scientific), according to the manufacturer’s instructions, on a ChemiDoc MP Imaging System (Bio-Rad).
Sample preparation for scRNA-seq
We generated scRNA-seq libraries using a Chromium Next GEM Single Cell 5′ kit (10x Genomics). Briefly, T cells labeled with TotalSeq Hashtag Antibody (BioLegend) from three biological replicates were sorted, which were then combined for each group. Following a wash step, cells were loaded onto a Chromium Single Cell Chip G to produce barcoded DNA. After DNA amplification, we prepared gene expression libraries using large cDNA fragments and cell surface protein libraries (HTO) from small DNA fragments (~200 bp). Gene expression and HTO libraries were multiplexed and sequenced on an Illumina NovaSeq 6000 using the same cycle settings described above.
Sample preparation for scATAC-seq + scRNA-seq
We generated scRNA-seq and scATAC-seq libraries using a Chromium Next GEM Single Cell Multiome ATAC + Gene Expression Reagent kit (10x Genomics), following the manufacturer’s user guide. Specifically, we sorted cells for nuclei isolation and followed the Low Cell Input Nuclei Isolation protocol from 10x Genomics to minimize nuclei loss. We tagmentated the isolated nuclei and captured them using the 10x Genomics Chromium Single Cell Controller. After GEM cleanup and preamplification PCR, we prepared gene expression and ATAC libraries separately. Sequencing of the gene expression libraries was performed on an Illumina NovaSeq 6000 using the following sequencing configuration: read 1 (26 cycles), i7 index (10 cycles), i5 index (10 cycles) and read 2 (90 cycles), targeting 30,000 reads per cell. The ATAC libraries were sequenced with read 1 (50 cycles), i7 index (8 cycles), i5 index (24 cycles) and read 2 (49 cycles), targeting 30,000 reads per cell.
Sample preparation for ChIP–seq
Ten million mouse CD19 CAR T cells transduced with pMIG or pMIG-BACH2 (human) were used to generate ChIP–seq libraries. A truChIP Chromatin Shearing kit (Covaris, 520154) was used for chromatin cross-linking, nuclei isolation and chromatin shearing. Ten micrograms of anti-BACH2 (Cell Signaling Technologies, 80775, clone D3T3G, 1:100) or anti-JunB (Cell Signaling Technologies, 3753, clone C37F9, 1:100) was used to pull down sheared chromatin from CD8+ T cells. ChIP–seq libraries were constructed using a published ChIPmentation protocol62. We sequenced the ChIP–seq libraries on an Illumina NextSeq 550, generating at least 20 million reads per sample.
Data analysis for scRNA-seq
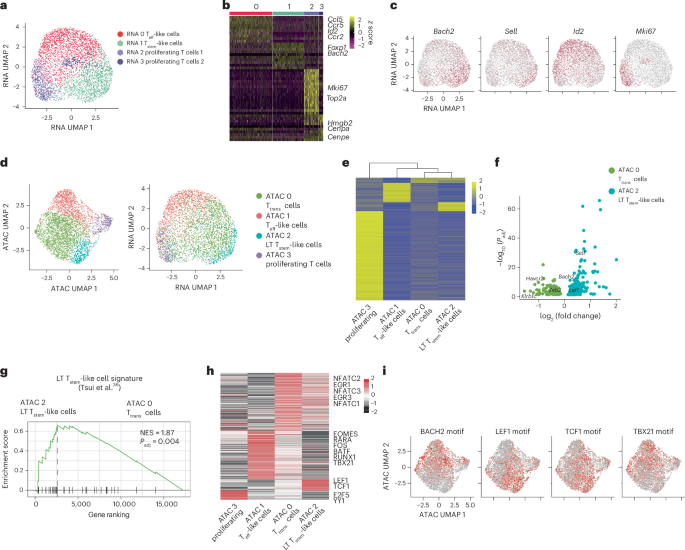
For human CAR T cell samples, the unique molecular identifier (UMI) count matrix for each sample was downloaded from the GSE241783 dataset. We used R package Seurat (v4.4.0) for the subsequent analysis. We merged cells from 40 individuals and filtered the cells with the following criteria: 200–7,000 detected genes, 1,000–15,000 detected RNA molecules and less than 15% mitochondrial genes. To focus on CD8+ CAR T cells, we selected CAR T cells expressing either CD8A or CD8B and not expressing CD4. Monocle 3 (ref. 63) was used to reconstruct developmental trajectories and infer transcriptional dynamics. The preprocessed Seurat object was imported into Monocle 3 while preserving the original UMAP embedding. Developmental lineages were inferred by applying the ‘learn_graph’ function, which fits a principal graph onto the UMAP manifold to capture the underlying cellular topology. Cells were then ordered along pseudotime using ‘order_cells’, with biologically defined root populations designated as starting points.
For mouse CAR T cell scRNA-seq fastq files, we used Cell Ranger (v6.0.0) to align FASTQ files to the mm10 reference genome and to quantify barcodes and UMIs for both the gene expression and hashtag libraries with the cellranger count pipeline. We combined the cells from different samples using the merge function and retained those with 1,000–3,500 detected genes, 2,000–15,000 detected RNA molecules and less than 5% mitochondrial gene content.
After quality control, we log normalized the data using a scale factor of 10,000. During the ScaleData function, we regressed out the effects of cell cycle, the number of detected genes and mitochondrial gene content. T cell antigen receptor and immunoglobulin genes were excluded from the top 2,000 variable genes identified with FindVariableFeatures, and the remaining genes were used for principal component analysis via RunPCA. We used the top 20 principal components to calculate neighbors, cluster the cell types and generate the UMAPs. We used the FindAllMarkers function or FindMarkers (min.pct = 0.1, logfc.threshold = 0.1) to determine markers genes in each cluster and the HTODemux function to determine Hashtag antibody labeling of each biological replicate. We used pheatmap (v1.0.12) or the DoHeatmap function to generate heat maps. We calculated gene set enrichment at the single-cell level using the AddModuleScore function. Additionally, we performed GSEA for two clusters of cells using the clusterProfiler package (v4.12.6).
Data analysis for scATAC-seq + scRNA-seq
scATAC-seq + scRNA-seq data were analyzed as previously described16. Cell versus gene matrices of UMI counts for gene expression assays and cell versus fragment matrixes for ATAC assays were generated from fastq files aligned to mm10 using cellranger-arc (v2.0.0). We performed subsequent analyses using Signac (v1) and Seurat (v4). Cells were retained if they had 1,000–4,000 detected genes, 15,000–40,000 detected ATAC fragments and less than 8% UMIs mapped to the mitochondrial genome. Additionally, we excluded cells with fewer than 50% of ATAC reads mapped to peaks, a nucleosome signal greater than 2 and a transcription start site enrichment score below 2. Using the approach outlined in ‘Data analysis for scRNA-seq’, we analyzed the scRNA-seq data to calculate UMAPs and identify clusters based on gene expression. For the scATAC-seq data, peaks were called using MACS2, and those located in the mm10 genomic blacklist or nonstandard chromosomes were removed. The FeatureMatrix function was used to quantify counts within the peaks, generating a peak assay for downstream analyses. After normalizing the data, top variable features were identified using FindTopFeatures. Latent semantic indexing was performed using RunSVD, and components 2 through 30 were used to calculate UMAPs and identify clusters. We used the geneactivity function to generate RNA activity assays and the chromVAR function to generate the motif assay. Differentially accessible peaks, RNA activity and motif activity were identified using FindAllMarkers or FindMarkers. We used the AddModuleScore function to calculate enrichment of the open chromatin signature of Asxl1-KO stem-like T cells. The number of correlated genes was determined using the dorcJPlot function from FigR (v0.1.0).
Data analysis for ChIP–seq
We analyzed the ChIP–seq data as previously described14. We mapped the ChIP–seq reads to the mouse genome (mm10) with Bowtie 1.1.1. We called peaks with MACS (v1.4.2; default P value threshold of 1 × 10−4). We used HOMER version 4.9 for peak annotation and transcription factor motif enrichment. We visualized transcription factor binding profiles using Integrative Genomics Viewer.
Statistical analysis
For statistical analysis, Prism (GraphPad, v10.4.0) and R (v4.1.3) were used. A two-tailed Student’s t-test was used for calculating the statistical significance between two experimental conditions. Data distribution was assumed to be normal, but this was not formally tested. A one-way or two-way ANOVA was used for determining statistical significance among more than two experimental groups. A P value of <0.05 was considered statistically significant. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications16,38,39. Mice used for all experiments were randomly assigned to each group. Data collection and analysis were not performed blind to the conditions of the experiments. No data points were excluded from the statistical analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.