Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63. https://doi.org/10.1038/nature08909.
Gilbert W. Why genes in pieces? Nature. 1978;271:501–501. https://doi.org/10.1038/271501a0.
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. https://doi.org/10.1038/ng.259.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. https://doi.org/10.1038/nature07509.
Marasco LE, Kornblihtt AR. The physiology of alternative splicing. Nat Rev Mol Cell Biol. 2023;24:242–54. https://doi.org/10.1038/s41580-022-00545-z.
Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–4. https://doi.org/10.1126/science.aad9417.
Gotthardt M, Badillo-Lisakowski V, Parikh VN, Ashley E, Furtado M, Carmo-Fonseca M, et al. Cardiac splicing as a diagnostic and therapeutic target. Nat Rev Cardiol. 2023;20:517–30. https://doi.org/10.1038/s41569-022-00828-0.
Bhattoa HP, Konstantynowicz J, Laszcz N, Wojcik M, Pludowski P. Vitamin D. Musculoskeletal health. Rev Endocr Metab Disord. 2017;18:363–71. https://doi.org/10.1007/s11154-016-9404-x.
Hossein-nezhad A, Holick MF. Vitamin D for health: a global perspective. Mayo Clin Proc. 2013;88:720–55. https://doi.org/10.1016/j.mayocp.2013.05.011.
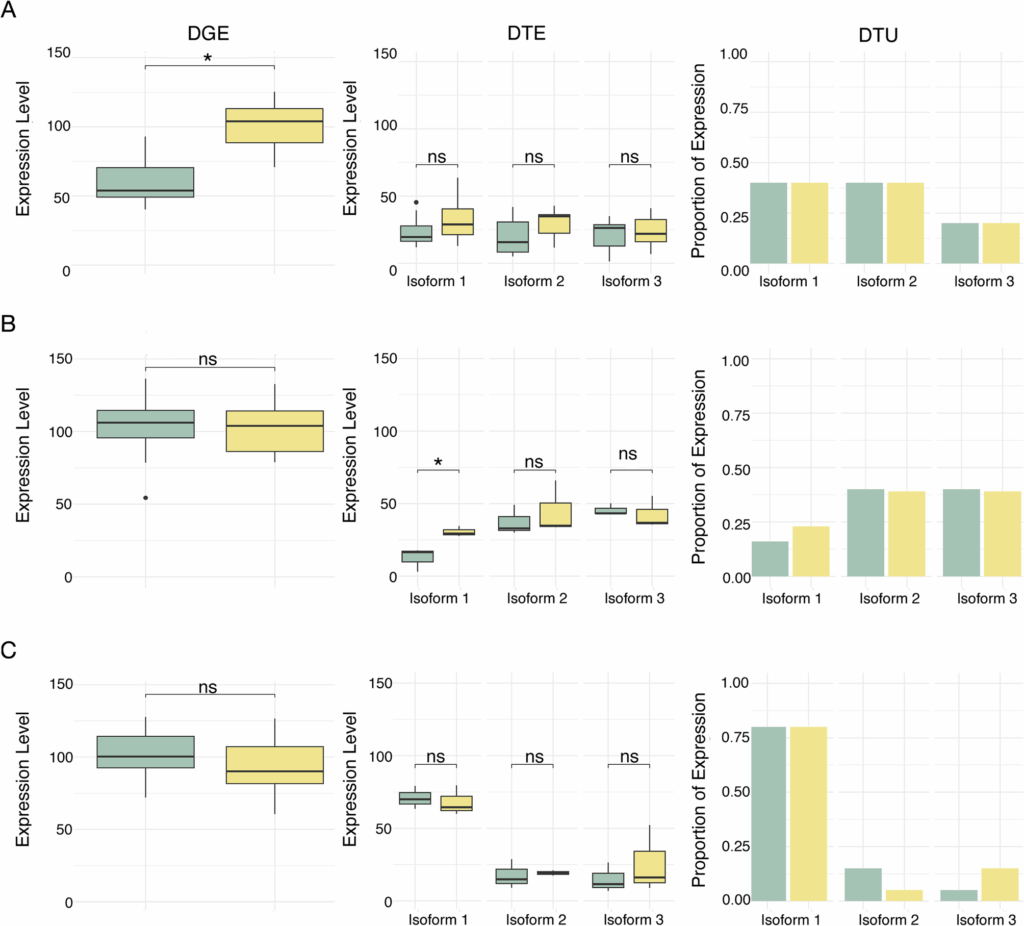
Gorman CE, Egan F, Alarcón-López FJ, Jakobsen J, McGinnity P, Hulsey CD. Vitamin D modulates gene expression in four major muscle tissues in Atlantic salmon. 2025. https://doi.org/10.21203/rs.3.rs-5267662/v1.
Tress ML, Abascal F, Valencia A. Alternative splicing may not be the key to proteome complexity. Trends Biochem Sci. 2017;42:98–110. https://doi.org/10.1016/j.tibs.2016.08.008.
Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. https://doi.org/10.1152/physrev.1996.76.2.371.
Henderson CA, Gomez CG, Novak SM, Mi-Mi L, Gregorio CC. Overview of the muscle cytoskeleton. Compr Physiol. 2017;7:891–944. https://doi.org/10.1002/cphy.c160033.
Bottinelli R. Functional heterogeneity of mammalian single muscle fibres: do myosin isoforms tell the whole story? Pflugers Arch. 2001;443:6–17. https://doi.org/10.1007/s004240100700.
Li A, Nelson SR, Rahmanseresht S, Braet F, Cornachione AS, Previs SB, et al. Skeletal MyBP-C isoforms tune the molecular contractility of divergent skeletal muscle systems. Proc Natl Acad Sci U S A. 2019;116:21882–92. https://doi.org/10.1073/pnas.1910549116.
Arora AS, Huang H-L, Singh R, Narui Y, Suchenko A, Hatano T, et al. Structural insights into actin isoforms. eLife. 2023;12:e82015. https://doi.org/10.7554/eLife.82015.
Gogulothu R, Nagar D, Gopalakrishnan S, Garlapati VR, Kallamadi PR, Ismail A. Disrupted expression of genes essential for skeletal muscle fibre integrity and energy metabolism in vitamin D deficient rats. J Steroid Biochem Mol Biol. 2020;197:105525. https://doi.org/10.1016/j.jsbmb.2019.105525.
Dirks-Naylor AJ, Lennon-Edwards S. The effects of vitamin D on skeletal muscle function and cellular signaling. J Steroid Biochem Mol Biol. 2011;125:159–68. https://doi.org/10.1016/j.jsbmb.2011.03.003.
Latham CM, Brightwell CR, Keeble AR, Munson BD, Thomas NT, Zagzoog AM, et al. Vitamin d promotes skeletal muscle regeneration and mitochondrial health. Front Physiol. 2021. https://doi.org/10.3389/fphys.2021.660498.
Endo I, Inoue D, Mitsui T, Umaki Y, Akaike M, Yoshizawa T, et al. Deletion of vitamin D receptor gene in mice results in abnormal skeletal muscle development with deregulated expression of myoregulatory transcription factors. Endocrinology. 2003;144:5138–44. https://doi.org/10.1210/en.2003-0502.
Zhou R, Chun RF, Lisse TS, Garcia AJ, Xu J, Adams JS, et al. Vitamin D and alternative splicing of RNA. J Steroid Biochem Mol Biol. 2015;148:310–7. https://doi.org/10.1016/j.jsbmb.2014.09.025.
Bischoff-Ferrari H, Borchers M, Gudat F, Dürmüller U, Stähelin H, Dick W. Vitamin D receptor expression in human muscle tissue decreases with age. J Bone Miner Res. 2004;19:265–9. https://doi.org/10.1359/jbmr.2004.19.2.265.
Pfeifer M, Begerow B, Minne HW. Vitamin D and muscle function. Osteoporos Int. 2002;13:187–94. https://doi.org/10.1007/s001980200012.
Ceglia L, Harris SS. Vitamin D and its role in skeletal muscle. Calcif Tissue Int. 2013;92:151–62. https://doi.org/10.1007/s00223-012-9645-y.
Jacobs A, Elmer KR. Alternative splicing and gene expression play contrasting roles in the parallel phenotypic evolution of a salmonid fish. Mol Ecol. 2021;30:4955–69. https://doi.org/10.1111/mec.15817.
Verta J-P, Debes PV, Piavchenko N, Ruokolainen A, Ovaskainen O, Moustakas-Verho JE, et al. Cis-regulatory differences in isoform expression associate with life history strategy variation in Atlantic salmon. PLoS Genet. 2020;16:e1009055. https://doi.org/10.1371/journal.pgen.1009055.
Jakobsen J, Smith C, Bysted A, Cashman KD. Vitamin D in wild and farmed Atlantic salmon (Salmo salar)—what do we know? Nutrients. 2019;11:982. https://doi.org/10.3390/nu11050982.
Atsuko T, Toshio O, Makoto T, Tadashi K. Possible origin of extremely high contents of vitamin D3 in some kinds of fish liver. Comparative Biochemistry and Physiology Part A: Physiology. 1991;100:483–7. https://doi.org/10.1016/0300-9629(91)90504-6.
Lock E-J, Waagbø R, Wendelaar Bonga S, Flik G. The significance of vitamin D for fish: a review. Aquac Nutr. 2010;16:100–16. https://doi.org/10.1111/j.1365-2095.2009.00722.x.
Choi YM, Kim BC. Muscle fiber characteristics, myofibrillar protein isoforms, and meat quality. Livest Sci. 2009;122:105–18. https://doi.org/10.1016/j.livsci.2008.08.015.
Noto RE, Leavitt L, Edens MA. Physiology, muscle. StatPearls. Treasure Island. (FL): StatPearls Publishing; 2024.
Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12:349–61. https://doi.org/10.1038/nrm3118.
Shih HP, Gross MK, Kioussi C. Muscle development: forming the head and trunk muscles. Acta Histochem. 2008;110:97–108. https://doi.org/10.1016/j.acthis.2007.08.004.
Chang C-N, Kioussi C, Location. Location, location: signals in muscle specification. J Dev Biol. 2018;6:11. https://doi.org/10.3390/jdb6020011.
Ripa R, George T, Shumway KR, Sattar Y, Physiology. In: StatPearls, editor. Cardiac muscle. Treasure Island (FL): StatPearls Publishing; 2024.
Steg A, Oczkowicz M, Świątkiewicz M. Effects of high-dose vitamin D3 supplementation on pig performance, vitamin D content in meat, and muscle transcriptome in pigs. J Anim Physiol Anim Nutr. 2025;109:560–73. https://doi.org/10.1111/jpn.14066.
Hangelbroek RWJ, Vaes AMM, Boekschoten MV, Verdijk LB, Hooiveld GJEJ, van Loon LJC, et al. No effect of 25-hydroxyvitamin D supplementation on the skeletal muscle transcriptome in vitamin D deficient frail older adults. BMC Geriatr. 2019;19:151. https://doi.org/10.1186/s12877-019-1156-5.
Yi L, Pimentel H, Bray NL, Pachter L. Gene-level differential analysis at transcript-level resolution. Genome Biol. 2018;19:53. https://doi.org/10.1186/s13059-018-1419-z.
Nowicka M, Robinson MD. DRIMSeq: a Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Res. 2016;5:1356. https://doi.org/10.12688/f1000research.8900.2.
Love MI, Soneson C, Patro R. Swimming downstream: statistical analysis of differential transcript usage following salmon quantification. F1000Res. 2018;7:952. https://doi.org/10.12688/f1000research.15398.3.
Wang Y, Xie Z, Kutschera E, Adams JI, Kadash-Edmondson KE, Xing Y. RMATS-turbo: an efficient and flexible computational tool for alternative splicing analysis of large-scale RNA-seq data. Nat Protoc. 2024;19:1083–104. https://doi.org/10.1038/s41596-023-00944-2.
Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437–51. https://doi.org/10.1038/nrm.2017.27.
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–9. https://doi.org/10.1038/nmeth.4197.
Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2016. https://doi.org/10.12688/f1000research.7563.2.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. https://doi.org/10.1186/s13059-014-0550-8.
Van den Berge K, Soneson C, Robinson MD, Clement L. StageR: a general stage-wise method for controlling the gene-level false discovery rate in differential expression and differential transcript usage. Genome Biol. 2017;18:151. https://doi.org/10.1186/s13059-017-1277-0.
Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38:276–8. https://doi.org/10.1038/s41587-020-0439-x.
Grüning B, Dale R, Sjödin A, Chapman BA, Rowe J, Tomkins-Tinch CH, et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods. 2018;15:475–6. https://doi.org/10.1038/s41592-018-0046-7.
da Veiga Leprevost F, Grüning BA, Alves Aflitos S, Röst HL, Uszkoreit J, Barsnes H, et al. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics. 2017;33:2580–2. https://doi.org/10.1093/bioinformatics/btx192.
Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35:316–9. https://doi.org/10.1038/nbt.3820.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. https://doi.org/10.14806/ej.17.1.200.
Andrews S. FastQC a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 16 May 2025.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. https://doi.org/10.1093/bioinformatics/bts635.
Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. https://doi.org/10.12688/f1000research.7563.2.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B: Statistical Methodology. 1995;57:289–300.
Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2018;35:2084–92. https://doi.org/10.1093/bioinformatics/bty895.
Pagès H, Carlson M, Aboyoun P, Falcon S, Morgan M. txdbmaker: Tools for making TxDb objects from genomic annotations. R package version 1.4.2. 2025. https://bioconductor.org/packages/txdbmaker; https://doi.org/10.18129/B9.bioc.txdbmaker.
Kolberg L, Raudvere U, Kuzmin I, Adler P, Vilo J, Peterson H. g:Profiler—interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023;51:W207–12. https://doi.org/10.1093/nar/gkad347.
Seguchi O, Takashima S, Yamazaki S, Asakura M, Asano Y, Shintani Y, et al. A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J Clin Invest. 2007;117:2812–24. https://doi.org/10.1172/JCI30804.
Warren SA, Briggs LE, Zeng H, Chuang J, Chang EI, Terada R, et al. Myosin light chain phosphorylation is critical for adaptation to cardiac stress. Circulation. 2012;126:2575–88. https://doi.org/10.1161/CIRCULATIONAHA.112.116202.
Davis JS, Hassanzadeh S, Winitsky S, Lin H, Satorius C, Vemuri R, et al. The overall pattern of cardiac contraction depends on a spatial gradient of myosin regulatory light chain phosphorylation. Cell. 2001;107:631–41. https://doi.org/10.1016/S0092-8674(01)00586-4.
Moss RL, Fitzsimons DP. Myosin light chain 2 into the mainstream of cardiac development and contractility. Circ Res. 2006;99:225–7. https://doi.org/10.1161/01.RES.0000236793.88131.dc.
Nawata J, Ohno I, Isoyama S, Suzuki J, Miura S, Ikeda J, et al. Differential expression of α1, α3 and α5 integrin subunits in acute and chronic stages of myocardial infarction in rats. Cardiovasc Res. 1999;43:371–81. https://doi.org/10.1016/S0008-6363(99)00117-0.
Neiman G, Scarafía MA, La Greca A, Santín Velazque NL, Garate X, Waisman A, et al. Integrin alpha-5 subunit is critical for the early stages of human pluripotent stem cell cardiac differentiation. Sci Rep. 2019;9:18077. https://doi.org/10.1038/s41598-019-54352-2.
Schumacher JA, Wright ZA, Owen ML, Bredemeier NO, Sumanas S. Integrin α5 and integrin α4 cooperate to promote endocardial differentiation and heart morphogenesis. Dev Biol. 2020;465:46–57. https://doi.org/10.1016/j.ydbio.2020.06.006.
Terracio L, Rubin K, Gullberg D, Balog E, Carver W, Jyring R, et al. Expression of collagen binding integrins during cardiac development and hypertrophy. Circ Res. 1991;68:734–44. https://doi.org/10.1161/01.res.68.3.734.
Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–31. https://doi.org/10.1093/nar/gkl924.
Wang Y, Liu J, Huang B, Xu Y-M, Li J, Huang L-F, et al. Mechanism of alternative splicing and its regulation. Biomed Rep. 2014;3:152–8. https://doi.org/10.3892/br.2014.407.
Girgis CM, Clifton-Bligh RJ, Hamrick MW, Holick MF, Gunton JE. The roles of vitamin D in skeletal muscle: form, function, and metabolism. Endocr Rev. 2013;34:33–83. https://doi.org/10.1210/er.2012-1012.
Gunton JE, Girgis CM. Vitamin D and muscle. Bone Rep. 2018;8:163–7. https://doi.org/10.1016/j.bonr.2018.04.004.
de la Guía-Galipienso F, Martínez-Ferran M, Vallecillo N, Lavie CJ, Sanchis-Gomar F, Pareja-Galeano H. Vitamin D and cardiovascular health. Clin Nutr. 2021;40:2946–57. https://doi.org/10.1016/j.clnu.2020.12.025.
den Berge KV, Hembach KM, Soneson C, Tiberi S, Clement L, Love MI et al. RNA Sequencing Data: Hitchhiker’s Guide to Expression Analysis. Annu Rev of Biomed Data Sci. 2019;2 Volume 2, 2019:139–73. https://doi.org/10.1146/annurev-biodatasci-072018-021255.
Bhuiyan SA, Ly S, Phan M, Huntington B, Hogan E, Liu CC, et al. Systematic evaluation of isoform function in literature reports of alternative splicing. BMC Genomics. 2018;19:637. https://doi.org/10.1186/s12864-018-5013-2.
Wright CJ, Smith CWJ, Jiggins CD. Alternative splicing as a source of phenotypic diversity. Nat Rev Genet. 2022;23:697–710. https://doi.org/10.1038/s41576-022-00514-4.
Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, et al. Function of alternative splicing. Gene. 2005;344:1–20. https://doi.org/10.1016/j.gene.2004.10.022.
Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, et al. Function of alternative splicing. Gene. 2013;514:1–30. https://doi.org/10.1016/j.gene.2012.07.083.
Garg A, Jansen S, Greenberg L, Zhang R, Lavine KJ, Greenberg MJ. Dilated cardiomyopathy-associated skeletal muscle actin (ACTA1) mutation R256H disrupts actin structure and function and causes cardiomyocyte hypocontractility. Proc Natl Acad Sci U S A. 2024;121:e2405020121. https://doi.org/10.1073/pnas.2405020121.