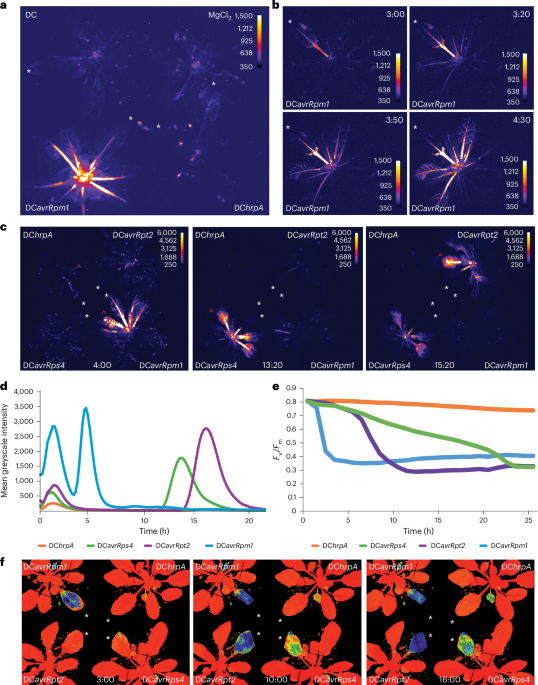
Cui, J. et al. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl Acad. Sci. USA 102, 1791–1796 (2005).
Shine, M. B., Xiao, X., Kachroo, P. & Kachroo, A. Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 279, 81–86 (2019).
Riedlmeier, M. et al. Monoterpenes support systemic acquired resistance within and between plants. Plant Cell 29, 1440–1459 (2017).
Wang, C. et al. Extracellular pyridine nucleotides trigger plant systemic immunity through a lectin receptor kinase/BAK1 complex. Nat. Commun. 10, 4810 (2019).
Shine, M. et al. Phased small RNA–mediated systemic signaling in plants. Sci. Adv. 8, eabm8791 (2022).
Liu, H. et al. Piperideine-6-carboxylic acid regulates vitamin B6 homeostasis and modulates systemic immunity in plants. Nat. Plants 11, 263–278 (2025).
Wendehenne, D., Gao, Q. M., Kachroo, A. & Kachroo, P. Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20, 127–134 (2014).
Yu, K. et al. A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 3, 1266–1278 (2013).
Gao, Q. M. et al. Mono- and digalactosyldiacylglycerol lipids function nonredundantly to regulate systemic acquired resistance in plants. Cell Rep. 9, 1681–1691 (2014).
Kachroo, A. & Kachroo, P. Mobile signals in systemic acquired resistance. Curr. Opin. Plant Biol. 58, 41–47 (2020).
Wenig, M. et al. Systemic acquired resistance networks amplify airborne defense cues. Nat. Commun. 10, 3813 (2019).
Lim, G.-H. et al. The plant cuticle regulates apoplastic transport of salicylic acid during systemic acquired resistance. Sci. Adv. 6, eaaz0478 (2020).
Lim, G.-H. et al. Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe 19, 541–549 (2016).
Shah, J., Chaturvedi, R., Chowdhury, Z., Venables, B. & Petros, R. A. Signaling by small metabolites in systemic acquired resistance. Plant J. 79, 645–658 (2014).
Zeier, J. Metabolic regulation of systemic acquired resistance. Curr. Opin. Plant Biol. 62, 102050 (2021).
Vlot, A. C. et al. Systemic propagation of immunity in plants. N. Phytol. 229, 1234–1250 (2021).
Grant, M. R. et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846 (1995).
Grant, M. et al. The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23, 441–450 (2000).
Jacob, P. et al. Plant ‘helper’ immune receptors are Ca2+-permeable nonselective cation channels. Science 373, 420–425 (2021).
Bennett, M., Mehta, M. & Grant, M. Biophoton imaging: a nondestructive method for assaying R gene responses. Mol. Plant Microbe Interact. 18, 95–102 (2005).
Birtic, S. et al. Using spontaneous photon emission to image lipid oxidation patterns in plant tissues. Plant J. 67, 1103–1115 (2011).
Truman, W., Bennett, M. H., Kubigsteltig, I., Turnbull, C. & Grant, M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl Acad. Sci. USA 104, 1075–1080 (2007).
Kiep, V. et al. Systemic cytosolic Ca(2+) elevation is activated upon wounding and herbivory in Arabidopsis. N. Phytol. 207, 996–1004 (2015).
Kiefer, I. W. & Slusarenko, A. J. The pattern of systemic acquired resistance induction within the Arabidopsis rosette in relation to the pattern of translocation. Plant Physiol. 132, 840–847 (2003).
Bent, A. F. et al. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860 (1994).
Gassmann, W., Hinsch, M. E. & Staskawicz, B. J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR–NBS–LRR family of disease-resistance genes. Plant J. 20, 265–277 (1999).
Littlejohn, G. R., Breen, S., Smirnoff, N. & Grant, M. Chloroplast immunity illuminated. N. Phytol. https://doi.org/10.1111/nph.17076 (2020).
Fu, Z. Q. et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232 (2012).
Rate, D. N. & Greenberg, J. T. The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27, 203–211 (2001).
Cao, H., Bowling, S. A., Gordon, A. S. & Dong, X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592 (1994).
Liu, L. et al. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7, 13099 (2016).
Zheng, X. Y. et al. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11, 587–596 (2012).
Wildermuth, M. C., Dewdney, J., Wu, G. & Ausubel, F. M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565 (2001).
Hartmann, M. et al. Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173, 456–469 e416 (2018).
Thines, B. et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665 (2007).
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y. & Howe, G. A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl Acad. Sci. USA 105, 7100–7105 (2008).
Torres Zabala, M. et al. Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. N. Phytol. 209, 1120–1134 (2015).
Brooks, D. M. et al. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 17, 162–174 (2004).
Zhang, L., Zhang, F., Melotto, M., Yao, J. & He, S. Y. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 68, 1371–1385 (2017).
Mousavi, S. A., Chauvin, A., Pascaud, F., Kellenberger, S. & Farmer, E. E. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426 (2013).
Ellis, C. & Turner, J. A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215, 549–556 (2002).
Park, J. H. et al. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12 (2002).
Li, M., Yu, G., Cao, C. & Liu, P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2, 100231 (2021).
Wasternack, C. & Song, S. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321 (2017).
Cucurou, C., Battioni, J. P., Thang, D. C., Nam, N. H. & Mansuy, D. Mechanisms of inactivation of lipoxygenases by phenidone and BW755C. Biochemistry 30, 8964–8970 (1991).
Farmer, E. E., Caldelari, D., Pearce, G., Walker-Simmons, M. K. & Ryan, C. A. Diethyldithiocarbamic acid inhibits the octadecanoid signaling pathway for the wound induction of proteinase inhibitors in tomato leaves. Plant Physiol. 106, 337–342 (1994).
Meesters, C. et al. A chemical inhibitor of jasmonate signaling targets JAR1 in Arabidopsis thaliana. Nat. Chem. Biol. 10, 830–836 (2014).
Nguyen, C. T., Kurenda, A., Stolz, S., Chetelat, A. & Farmer, E. E. Identification of cell populations necessary for leaf-to-leaf electrical signaling in a wounded plant. Proc. Natl Acad. Sci. USA 115, 10178–10183 (2018).
Choi, W. G., Hilleary, R., Swanson, S. J., Kim, S. H. & Gilroy, S. Rapid, long-distance electrical and calcium signaling in plants. Annu. Rev. Plant Biol. 67, 287–307 (2016).
Toyota, M. et al. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112–1115 (2018).
Yan, C. et al. Ca(2+)/calmodulin-mediated desensitization of glutamate receptors shapes plant systemic wound signalling and anti-herbivore defence. Nat. Plants 10, 145–160 (2024).
Wang, J., Song, W. & Chai, J. Structure, biochemical function, and signaling mechanism of plant NLRs. Mol. Plant 16, 75–95 (2023).
de Torres-Zabala, M. et al. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants 1, 15074 (2015).
Mur, L. A., Kenton, P., Atzorn, R., Miersch, O. & Wasternack, C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262 (2006).
Jacob, P., Hige, J. & Dangl, J. L. Is localized acquired resistance the mechanism for effector-triggered disease resistance in plants? Nat. Plants https://doi.org/10.1038/s41477-023-01466-1 (2023).
Betsuyaku, S. et al. Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol. 59, 8–16 (2018).
Andersson, M. X. et al. Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana: formation of a novel oxo-phytodienoic acid-containing galactolipid, arabidopside E. J. Biol. Chem. 281, 31528–31537 (2006).
Zoeller, M. et al. Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiol. 160, 365–378 (2012).
Li, Q., Wang, C. & Mou, Z. Perception of damaged self in plants. Plant Physiol. 182, 1545–1565 (2020).
Vega-Munoz, I. et al. Breaking bad news: dynamic molecular mechanisms of wound response in plants. Front. Plant Sci. 11, 610445 (2020).
Yuan, M., Ngou, B. P. M., Ding, P. & Xin, X. F. PTI–ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030 (2021).
Bjornson, M., Pimprikar, P., Nurnberger, T. & Zipfel, C. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat. Plants 7, 579–586 (2021).
Manzoor, H. et al. Involvement of the glutamate receptor AtGLR3.3 in plant defense signaling and resistance to Hyaloperonospora arabidopsidis. Plant J. 76, 466–480 (2013).
Perkins, L. E. et al. Generalist insects behave in a jasmonate-dependent manner on their host plants, leaving induced areas quickly and staying longer on distant parts. Proc. R. Soc. B 280, 20122646 (2013).
Morin, H. et al. Wound-response jasmonate dynamics in the primary vasculature. N. Phytol. 240, 1484–1496 (2023).
Gilroy, S. et al. ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol. 171, 1606–1615 (2016).
de Torres, M. et al. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 47, 368–382 (2006).
Procko, C. et al. Leaf cell-specific and single-cell transcriptional profiling reveals a role for the palisade layer in UV light protection. Plant Cell 34, 3261–3279 (2022).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
O’Malley, R. C., Alonso, J. M., Kim, C. J., Leisse, T. J. & Ecker, J. R. An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2, 2910–2917 (2007).
Engler, C. et al. A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol. 3, 839–843 (2014).
King, E. O., Ward, M. K. & Raney, D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307 (1954).
Rufian, J. S., Rueda-Blanco, J., Beuzon, C. R. & Ruiz-Albert, J. Protocol: an improved method to quantify activation of systemic acquired resistance (SAR). Plant Methods 15, 16 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).