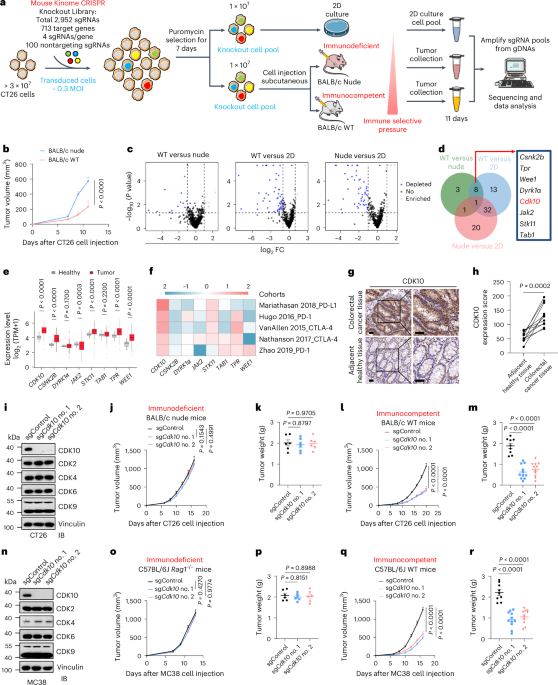
Zou, W., Wolchok, J. D. & Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8, 328rv324 (2016).
Sharma, P. et al. Immune checkpoint therapy-current perspectives and future directions. Cell 186, 1652–1669 (2023).
Iwasaki, A. & Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353 (2015).
Carroll, S. L., Pasare, C. & Barton, G. M. Control of adaptive immunity by pattern recognition receptors. Immunity 57, 632–648 (2024).
Demaria, O. et al. Harnessing innate immunity in cancer therapy. Nature 574, 45–56 (2019).
Cao, L. L. & Kagan, J. C. Targeting innate immune pathways for cancer immunotherapy. Immunity 56, 2206–2217 (2023).
Lynch, C., Pitroda, S. P. & Weichselbaum, R. R. Radiotherapy, immunity, and immune checkpoint inhibitors. Lancet Oncol. 25, e352–e362 (2024).
Kornepati, A. V. R., Rogers, C. M., Sung, P. T. & Curiel, T. J. The complementarity of DDR, nucleic acids and anti-tumour immunity. Nature 619, 475–486 (2023).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
Sheng, W. et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563 e519 (2018).
Maxwell, M. B. et al. ARID1A suppresses R-loop-mediated STING-type I interferon pathway activation of anti-tumor immunity. Cell 187, 3390–3408 e3319 (2024).
Zhang, Z. D. & Zhong, B. Regulation and function of the cGAS-MITA/STING axis in health and disease. Cell Insight 1, 100001 (2022).
Jiang, X. et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973 (2012).
Goel, S. et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017).
Deng, J. et al. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 8, 216–233 (2018).
Fassl, A., Geng, Y. & Sicinski, P. CDK4 and CDK6 kinases: from basic science to cancer therapy. Science 375, eabc1495 (2022).
Alvarez-Fernandez, M. & Malumbres, M. Mechanisms of sensitivity and resistance to CDK4/6 Inhibition. Cancer Cell 37, 514–529 (2020).
Dietrich, C. et al. INX-315, a selective CDK2 inhibitor, induces cell cycle arrest and senescence in solid tumors. Cancer Discov. 14, 446–467 (2024).
Arora, M. et al. Rapid adaptation to CDK2 inhibition exposes intrinsic cell-cycle plasticity. Cell 186, 2628–2643 e2621 (2023).
Cidado, J. et al. AZD4573 is a highly selective CDK9 inhibitor that suppresses MCL-1 and induces apoptosis in hematologic cancer cells. Clin. Cancer Res. 26, 922–934 (2020).
Parua, P. K. et al. A Cdk9-PP1 switch regulates the elongation-termination transition of RNA polymerase II. Nature 558, 460–464 (2018).
Hluchy, M. et al. CDK11 regulates pre-mRNA splicing by phosphorylation of SF3B1. Nature 609, 829–834 (2022).
Dubbury, S. J., Boutz, P. L. & Sharp, P. A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 564, 141–145 (2018).
Iniguez, A. B. et al. EWS/FLI confers tumor cell synthetic lethality to CDK12 inhibition in Ewing sarcoma. Cancer Cell 33, 202–216 e206 (2018).
Quereda, V. et al. Therapeutic targeting of CDK12/CDK13 in triple-negative breast cancer. Cancer Cell 36, 545–558 e547 (2019).
Insco, M. L. et al. Oncogenic CDK13 mutations impede nuclear RNA surveillance. Science 380, eabn7625 (2023).
Guen, V. J. et al. CDK10/cyclin M is a protein kinase that controls ETS2 degradation and is deficient in STAR syndrome. Proc. Natl Acad. Sci. USA 110, 19525–19530 (2013).
Iorns, E. et al. Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell 13, 91–104 (2008).
You, Y. et al. Downregulated CDK10 expression in gastric cancer: association with tumor progression and poor prognosis. Mol. Med. Rep. 17, 6812–6818 (2018).
Li, H., You, Y. & Liu, J. Cyclin‑dependent kinase 10 prevents glioma metastasis via modulation of Snail expression. Mol. Med. Rep. 18, 1165–1170 (2018).
Weiswald, L. B. et al. Inactivation of the kinase domain of CDK10 prevents tumor growth in a preclinical model of colorectal cancer, and is accompanied by downregulation of Bcl-2. Mol. Cancer Ther. 16, 2292–2303 (2017).
Windpassinger, C. et al. CDK10 mutations in humans and mice cause severe growth retardation, spine malformations, and developmental delays. Am. J. Hum. Genet. 101, 391–403 (2017).
Guen, V. J. et al. A homozygous deleterious CDK10 mutation in a patient with agenesis of corpus callosum, retinopathy, and deafness. Am. J. Med. Genet. A 176, 92–98 (2018).
Gu, S. S. et al. Therapeutically increasing MHC-I expression potentiates immune checkpoint blockade. Cancer Discov. 11, 1524–1541 (2021).
Manguso, R. T. et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017).
Belnap, L. P., Cleveland, P. H., Colmerauer, M. E., Barone, R. M. & Pilch, Y. H. Immunogenicity of chemically induced murine colon cancers. Cancer Res. 39, 1174–1179 (1979).
Wang, X. et al. In vivo CRISPR screens identify the E3 ligase Cop1 as a modulator of macrophage infiltration and cancer immunotherapy target. Cell 184, 5357–5374 e5322 (2021).
Guo, E. et al. WEE1 inhibition induces anti-tumor immunity by activating ERV and the dsRNA pathway. J. Exp. Med. https://doi.org/10.1084/jem.20210789 (2022).
Bell, H. N. & Zou, W. Beyond the barrier: unraveling the mechanisms of immunotherapy resistance. Annu. Rev. Immunol. 42, 521–550 (2024).
Della Corte, C. M. & Byers, L. A. Evading the STING: LKB1 loss leads to STING silencing and immune escape in KRAS-mutant lung cancers. Cancer Discov. 9, 16–18 (2019).
Zulato, E. et al. LKB1 loss is associated with glutathione deficiency under oxidative stress and sensitivity of cancer cells to cytotoxic drugs and γ-irradiation. Biochem. Pharmacol. 156, 479–490 (2018).
Skoulidis, F. et al. CTLA4 blockade abrogates KEAP1/STK11-related resistance to PD-(L)1 inhibitors. Nature 635, 462–471 (2024).
Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, 442–459 e429 (2020).
Dexter, D. L. et al. Heterogeneity of cancer cells from a single human colon carcinoma. Am. J. Med. 71, 949–956 (1981).
van den Heuvel, S. & Harlow, E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262, 2050–2054 (1993).
Robert, T. et al. Development of a CDK10/CycM in vitro kinase screening assay and identification of first small-molecule inhibitors. Front. Chem. 8, 147 (2020).
Brown, N. R., Noble, M. E., Endicott, J. A. & Johnson, L. N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1, 438–443 (1999).
Zhou, X. et al. Pharmacologic activation of p53 triggers viral mimicry response thereby abolishing tumor immune evasion and promoting antitumor immunity. Cancer Discov. 11, 3090–3105 (2021).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Crossley, M. P. et al. R-loop-derived cytoplasmic RNA-DNA hybrids activate an immune response. Nature 613, 187–194 (2023).
Yasuhara, T. et al. RAP80 suppresses the vulnerability of R-loops during DNA double-strand break repair. Cell Rep. 38, 110335 (2022).
Valk, E., Ord, M., Faustova, I. & Loog, M. CDK signaling via nonconventional CDK phosphorylation sites. Mol. Biol. Cell 34, pe5 (2023).
Kusubata, M. et al. cdc2 kinase phosphorylation of desmin at three serine/threonine residues in the amino-terminal head domain. Biochem. Biophys. Res. Commun. 190, 927–934 (1993).
Li, Z. et al. Synthesis and structure-activity relationships of cyclin-dependent kinase 11 inhibitors based on a diaminothiazole scaffold. Eur. J. Med. Chem. 238, 114433 (2022).
Duster, R., Ji, Y., Pan, K. T., Urlaub, H. & Geyer, M. Functional characterization of the human Cdk10/Cyclin Q complex. Open Biol. 12, 210381 (2022).
Sachs, R. E., Ginsburg, P. B. & Goldman, D. P. Encouraging new uses for old drugs. JAMA 318, 2421–2422 (2017).
Weisberg, E. et al. Antileukemic effects of the novel, mutant FLT3 inhibitor NVP-AST487: effects on PKC412-sensitive and -resistant FLT3-expressing cells. Blood 112, 5161–5170 (2008).
Kantarjian, H. M. et al. ponatinib-review of historical development, current status, and future research. Am. J. Hematol. 99, 1576–1585 (2024).
Eide, C. A. et al. Overcoming clinical BCR-ABL1 compound mutant resistance with combined ponatinib and asciminib therapy. Cancer Cell 42, 1486–1488 (2024).
Li, H. et al. USP8-governed GPX4 homeostasis orchestrates ferroptosis and cancer immunotherapy. Proc. Natl Acad. Sci. USA 121, e2315541121 (2024).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Dai, P. et al. USP2 inhibition unleashes CD47-restrained phagocytosis and enhances anti-tumor immunity. Nat. Commun. 16, 4564 (2025).
Crossley, M. P. et al. Catalytically inactive, purified RNase H1: a specific and sensitive probe for RNA-DNA hybrid imaging. J. Cell Biol. https://doi.org/10.1083/jcb.202101092 (2021).
Zhang, J. et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018).
Mariathasan, S. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018).
Zhao, J. et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 25, 462–469 (2019).
Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016).
Van Allen, E. M. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015).
Nathanson, T. et al. Somatic mutations and neoepitope homology in melanomas treated with CTLA-4 blockade. Cancer Immunol. Res. 5, 84–91 (2017).
Gong, L. et al. Cancer immunology data engine reveals secreted AOAH as a potential immunotherapy. Cell 188, 5062–5080 e5032 (2025).
Patil, N. S. et al. Intratumoral plasma cells predict outcomes to PD-L1 blockade in non-small cell lung cancer. Cancer Cell 40, 289–300 e284 (2022).