Study design
The aim of this study was to explore a target in T cells that suppressing allograft rejection while maintaining antitumor immunity. The goal was achieved by using Setdb1f/fCd4-Cre mice; a cardiac allograft rejection model; a chronic rejection model; and tumor models. For mechanistic study, we compared allograft survival time between Setdb1f/fCd4-Cre mice and Setdb1f/fFoxp3-Cre mice. Also, we performed several biochemistry experiments, such as bulk and single-cell RNA-seq, CUT&Tag assay, co-IP assay, flow cytometric analysis, immunofluorescence, quantitative real-time reverse transcription PCR (RTPCR) analysis, ex vivo Treg cell induction, etc. All data points generated were included in the analysis. For in vitro and in vivo studies, as least three independent experiments were performed. Besides, for in vivo studies, mice were randomly assigned for experiments. The sample size is specified in each figure legend. The experiments were performed unblinded. The key resources of this research were listed in Supplementary Data 4.
Mice
Specific pathogen-free C57BL/6JNifdc (stock number: 219) and BALB/c mice (stock number: 211) were purchased from Charles River Laboratories (Beijing, China).Cd4-Cre transgenic mice (stock number: 022071) were purchased from Model Organisms (Shanghai, China), and genotyped. Foxp3-GFP mice [C57BL/6-Tg(Foxp3-GFP)90Pkraj/J, stock number: 023800] and Foxp3-Cre mice [B6129S-Tg(Foxp3-EGFP/icre)1aJbs/J, stock number: 023161] were obtained from Jackson Laboratory (Bar Harbor, ME, USA). The Setdb1f/f mice were generated by the CRISPR-Cas9 system (GemPharmatech), and were backcrossed with Cd4-Cre transgenic mice or Foxp3-Cre mice, to generate mice with selective deletion of Setdb1 in T cells (Setdb1f/fCd4-Cre) or in Tregs (Setdb1f/fFoxp3-Cre). To generate the floxed allele, two gRNAs (gRNA1: TGCGTCCGACGTTTTACCTC, gRNA2: ATGAAGCCTGCAGGAACTAA) targeting introns flanking the critical exon(s) were used, and a donor vector containing loxP sites was introduced for homology-directed repair. The Cas9 protein, gRNAs, and donor DNA were microinjected into C57BL/6 zygotes. Positive founders were confirmed by PCR and sequencing. All mice were maintained under controlled conditions (22 °C, 50% humidity, 12 h light/dark cycle, with lights on at 7:00 AM). Mice were euthanized by CO2 inhalation followed by cervical dislocation, in accordance with institutional animal care guidelines.
In vivo cell depletion
T cell or NK cell was depleted by administering 200 μg of depleting antibodies i.p. weekly beginning three days prior to tumor implantation as indicated: CD4+ T cells with anti-CD4 (clone GK1.5, BioXCell), CD8+ T cells with anti-CD8α (clone 2.43, BioXCell), NK cells with anti-NK1.1 (clone PK136, BioXCell). For CD25+ cell depletion, purified CD25 antibody was administrated 100 μg per mice i.p. twice weekly, beginning three days prior to transplantation.
Transplantation procedures
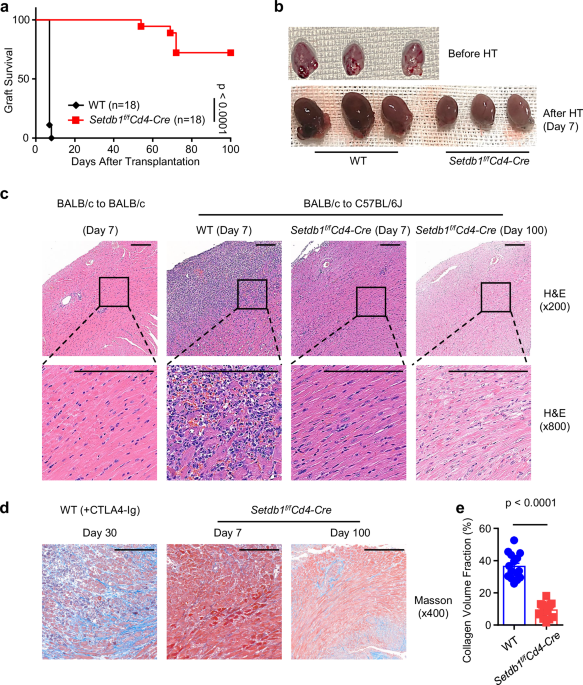
Heart transplantation in mice was performed according to a previously described method44. In brief, hearts from BALB/c donors were transplanted into 8- to 10-week-old male WT B6 or Setdb1f/fCd4-Cre recipient mice. The pulmonary artery and aorta of the donor heart were cut open, the remaining heart vessels were tied off, and the heart was removed. The chest cavity of an anesthetized recipient mouse was opened with a midline incision, and blood flow via the abdominal aorta and inferior vena cava was interrupted by ligation with 6-0 silk thread. Incisions were made in the recipient’s abdominal aorta and inferior vena cava to allow for anastomosis to be performed with the donor heart aorta and donor pulmonary artery, respectively. Anastomosis was made by 11-0 sutures running continuously; The 6-0 silk thread was removed, and the abdomen was then closed by 5-0 sutures in two layers. Heart graft survival was monitored daily by palpation, and the day of complete cessation of heartbeat was considered the day of rejection. Allografts were harvested for analysis at the time of rejection or at the indicated time points.
Skin transplantation in mice was performed as follows. BALB/c mice were euthanized, and their tails were swabbed with 70% ethanol. A 1.0 × 1.0 cm2 section of tail skin was removed from the donor. Recipient mice were anesthetized with isoflurane, and hair on their backs was removed with depilatory cream. The shaved area was cleaned with 70% ethanol and allowed to dry. A 1.0 × 1.0 cm2 area of back skin was removed from the recipient mice to create a graft bed. The donor skin graft was placed into the graft bed and stitched at the corners. The mice were then wrapped with bandages and placed in an incubator until they could move freely.
Histological examination and immunofluorescent staining
Grafts were fixed in 4% formaldehyde and dehydrated with xylene, absolute ethyl alcohol, and 75% alcohol, then embedded in paraffin wax. Four micron-thick sections were cut. Graft histological changes were revealed by hematoxylin & eosin (H&E) staining. Fibrosis degree was determined by Masson trichrome staining. T-cell infiltration into grafts was estimated by staining with anti-CD3 primary antibody according to a standard protocol (Servicebio).
For determination of p-ATF1/Setdb1 expression and intracellular co-localization in iTregs treated with or without p-ATF1 agonist (BC11-38), a cell pellet-based immunofluorescent staining was applied. In brief, cells were first fixed in 2% formaldehyde for 15 min and washed with PBS, and pellets were collected. A 2–4% liquid agarose was used for coating the pellets. After the agarose solidified, the clump was dehydrated with xylene, absolute ethyl alcohol, and 75% alcohol, then embedded in paraffin wax. Immunofluorescent staining was performed according to a standard protocol (LabEXTM).
Cell lines and tumor models
B16F1, Hepa1-6, and MC38 were purchased from the China Center for Type Culture Collection, and cultured in RPMI1640 medium (Gibco) plus 10% FBS (Yeasen Biotechnology Co., Ltd., Shanghai, China) and 1% penicillin/streptomycin (New Cell & Molecular Biotech Co., Ltd., Shanghai, China).
For the tumor model, 5 × 105 tumor cells were injected subcutaneously into the right flank of mice, and tumor growth was measured by multiplying the length of the tumor by the width. Tumors in mice were monitored continuously for 7 days starting on day 7 of tumor loading. For in vivo depletion T cells, anti-CD4 and anti-CD8 depletion antibodies or their corresponding isotype (BioXCell) were given at 200 μg per mouse at 3 days before tumor implant. To verify the effect of IL-18 on Treg cells and its anti-tumor efficacy, we transfected pCMV-Il18 or empty vector to B16F1, followed by puromycin screening, eventually obtaining the target cells.
Isolation of graft-infiltrating mononuclear cells, tumor-infiltrating mononuclear cells, splenocytes, and thymocytes
Mice bearing heart allografts were euthanized on day 7. Grafts were removed and cut into pieces, washed with PBS, and then digested at 37 °C for 30 min in DMEM containing 100 ng/mL type II collagenase (SigmalAdrich) before being pressed through a 70-μm filter (Biosharp). The collected cells were washed, and graft-infiltrating mononuclear cells were purified by density gradient centrifugation in a 38% Percoll solution (GE Healthcare).
For purification of tumor-infiltrating mononuclear cells, tumor-bearing mice were euthanized on day 14, and tumors were isolated from the surrounding tissue. Tumors were washed with PBS and then digested at 37 °C for 30 min in DMEM containing 100 ng/mL type IV collagenase (SigmalAdrich) before being pressed through a 70-μm filter (Biosharp). The collected cells were washed by PBS, followed by density gradient centrifugation in 38% Percoll solution (GE Healthcare).
Purified splenocytes were prepared by homogenization with a syringe, followed by passage through a 0.1-mm sterile nylon mesh and density gradient centrifugation in mouse 1× lymphocyte separation medium (Dakewe). Thymocytes were prepared by pressing thymus tissue through a 70-μm filter.
Flow cytometry
Cells were stained with various fluorochrome-conjugated antibodies, followed by analysis with a BD Celesta cell analyzer (BD Biosciences). Intracellular staining for Foxp3, T-bet, H3K4me3, H3K9me3, H3K27me3, and Setdb1 was performed using the Transcription Factor Buffer Set (BD Biosciences). Intracellular staining of IFN-γ, TNF-α, granzyme B, and perforin was performed according to the Intracellular Flow Cytometry Staining Protocol (Biolegend). For detection of intracellular IFN-γ and TNF-α, a cell-stimulation cocktail containing PMA/ionomycin and brefeldin A (eBioscience) was used to stimulate splenocytes for 4 h. Cytokine levels in mouse serum were measured with a CBA kit for mouse Th1/Th2/Th17 cytokines according to the manufacturer’s instructions. Data were analyzed with FlowJo software (TreeStar). Gating strategies have been provided in Supplementary Fig. 18.
Cell sorting
To induce Foxp3+ or T-bet+ T cells in vitro, naïve CD4+ T cells were sorted by magnetic bead separation by using Naive CD4+ T Cell Isolation Kit (Miltenyi). To assess the gene expression in thymocyte subsets, thymocytes from Foxp3-GFP reporter mice were depleted of CD8+ cells, followed by staining with PE-labeled anti-CD4 and APC-labeled anti-CD25 antibodies. T-cell subsets were sorted with a high-speed cell sorter FACSaria III (BD Biosciences).
In vitro T-cell stimulation
To activate T cells, naïve CD4+ T cells were added at 2 × 105 cells/well to anti-CD3 (5 μg/ml)-precoated 96-well round-bottom tissue culture plates (Corning); the culture medium contained 1 μg/ml soluble anti-CD28 mAb. For polarization of CD4+ T cells in vitro, cultures were supplemented with various cytokines (PeproTech, Rocky Hill, NJ) and small-molecule inhibitors (MCE). For Th1 polarization, naïve CD4+ cells were activated in the presence of 10 ng/ml mouse IL-2 and 10 ng/ml mouse IL-12. For Treg polarization, naïve CD4+ cells were activated in the presence of 10 ng/ml human TGF-β1 and 10 ng/ml mouse IL-2. For Th2 polarization, naïve CD4+ cells were activated in the presence of 10 ng/ml mouse IL-2 and 50 ng/ml mouse IL-4. For Th17 polarization, naïve CD4+ cells were activated in the presence of 50 ng/ml mouse IL-6 and 10 ng/ml human TGF-β. To promote ATF1 phosphorylation, BC11-38 was used at a concentration of 20 μM. CD4+ T cells cultured for different numbers of days were collected and analyzed by flow cytometry, immunoblotting, immunofluorescent staining, quantitative real-time PCR, co-immunoprecipitation, RNA-seq, and CUT&Tag.
Quantitative RT-PCR
Total RNA was extracted from T cells using a RNAfast200 kit (Fastagen), and cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara). The expression of genes of interest and of the HPRT control was assessed by PCR using SYBR Green mix (Yeasen). Transcript levels of target genes were calculated as the ratio of target gene expression to β-actin expression. Fold changes in target gene expression were analyzed by StepOne Software (Applied Biosystems) using the delta/delta CT method.
Immunoblot analysis
Protein extracts were resolved by SDS-PAGE, transferred onto an Immunobilon membrane, and analyzed by immunoblotting with the antibodies indicated. Horseradish peroxidase–linked antibody to rabbit immunoglobulin G (ThermoFisher) and horseradish peroxidase–linked antibody to mouse immunoglobulin G (ThermoFisher) were used as secondary antibodies. Protein expression was detected by chemiluminescence.
Co-immunoprecipitation assay
CD4+ T cells from WT mice were stimulated by anti-CD3/anti-CD28 for 72 h. Cells (4 × 106 cells) were collected in NETN lysis buffer. Subsequently, the cell lysate was incubated with anti-Setdb1 (1:50, CST) or control antibody at 4 °C for 2 h, followed by incubation with Magna ChIPTM Protein A + G Magnetic Beads (Merck) overnight. Beads were collected by centrifugation and washed 4 times with IP lysis buffer. Proteins were resolved in PBS and analyzed by immunoblotting.
Bulk RNA sequencing
Total RNA was extracted from purified CD4+ T cells with TRIzol (Invitrogen). Total RNAs (2 μg) were used for stranded RNA sequencing library preparation by means of a Stranded mRNA Library Prep Kit from DR08502 (Bioyigene) according to the manufacturer’s instructions. The library products corresponding to 200–500 bp were enriched, quantified, and finally sequenced on DNBSEQ-T7. The gene expression profiles of CD4+ T cells from WT B6 and Setdb1f/fCd4-Cre mice after various treatments were determined by RNA-Seq data analysis (Bioyigene). In brief, raw sequencing data were first filtered by FastQC; low-quality reads were discarded, and adaptor sequences were trimmed. After quality filtering, each sample had ~49.5–67.5 million clean reads. Clean reads from each sample were mapped to the Mus musculus GRCm38 reference genome using hisat2. Significantly differentially expressed transcripts were screened by applying the criteria FC ≥ 2 or ≤−2 and P-value < 0.05. The RNA-seq data, entitled “Transcriptome RNA-seq analysis for WT and Setdb1 cko mice CD4+ T cells before and after stimulation or transplantation” were deposited in the Sequence Read Archive (SRA) with BioProject number PRJNA843698.
Cleavage under targets and tagmentation (CUT&Tag)
CUT&Tag was performed using the Hyperactive Universal CUT&Tag Assay Kit. The sequencing process was outsourced to Novogene Co., Ltd. Briefly, after harvesting, CD25+ enriched T cells were counted and divided into groups of 200,000 cells each. The cells were centrifuged at 600g for 5 min at room temperature. Each sample was resuspended in 100 μl of pre-cooled NE Buffer, mixed gently, and incubated on ice. Following incubation, the cells were centrifuged again at 600g for 5 min at room temperature, and the pellets were resuspended in 100 μl of wash buffer.
Next, 10 μl of Concanavalin A-coated magnetic beads, pre-activated at room temperature, were added to each sample and incubated for 10 min. The unbound supernatant was removed, followed by an overnight reaction between the H3K9me3 group sample and anti-H3K9me3 antibodies (1:50, Proteintech), as well as between the SETDB1 group sample and anti-SETDB1 antibodies (2 μg, Proteintech) in Antibody Buffer at 4 °C.
After washing, the bound antibodies were reacted with secondary antibodies and then incubated with pA/G-Tnp Pro (0.04 μM). Next, the samples were fragmented with TTBL, followed by the addition of 10% SDS and DNA spike-in to each sample. To extract the genomic DNA, DNA Extract Beads Pro were activated and added to each sample. The extracted DNA fragments were amplified by PCR with indexing primers (Vazyme, TD202), and the library was purified using VAHTS DNA Clean Beads (Vazyme, N411).
Adapter sequences and low-quality reads in the raw CUT&Tag data were filtered using BBDuk (version 38.44). The filtered reads were then aligned to the mouse mm10 reference genome using Bowtie2 (version 2.4.2) with the parameters “–local –very-sensitive –no-mixed –no-discordant –phred33 -I 10 -X 700.” Duplicate reads were removed with the MarkDuplicates function in GATK (version 4.2.0). Coverage (bigWig) files were generated from the resulting BAM files using the bamCoverage tool in deepTools. Peak calling was performed using MACS2 (version 2.2.7.1) with the parameters “–keep-dup all –broad –broad-cutoff 0.05.” Finally, quantitative differences in binding and corresponding p-values were determined using MAnorm (version 1.3.0). Binding motifs were obtained using the findMotifsGenome.pl software in Homer (V5.1).
Single-cell RNA sequencing and data analysis
Thymus from 5 mice for each group were pooled together, isolated thymocytes, and roughly enriched for CD25+ cells using magnetic beads. The scRNA-seq libraries were generated using the 10× Genomics Chromium Controller Instrument and Chromium Single Cell 3′ V3 Reagent Kits (10× Genomics, Pleasanton, CA). Briefly, cells were concentrated to 1000 cells/µL and approximately 8000 cells were loaded into each channel to generate single-cell gel bead-in-emulsions (GEM), which results into expected mRNA barcoding of 5000 single-cells for each sample. After the RT step, GEMs were broken and barcoded-cDNA was purified and amplified. The amplified barcoded cDNA was fragmented, A-tailed, ligated with adaptors and index PCR amplified. The final libraries were quantified using the Qubit High Sensitivity DNA assay (Thermo Fisher Scientific) and the size distribution of the libraries were determined using a High Sensitivity DNA chip on a Bioanalyzer 2200 (Agilent). All libraries were sequenced by illumina sequencer (Illumina, San Diego, CA) on a 150 bp paired-end run.
scRNA-seq data analysis for CD4+ T cells was performed with NovelBrain Cloud Analysis Platform (www.novelbrain.com). Seurat package (version: 4.0.3, https://satijalab.org/seurat/) was used for cell normalization and regression based on the expression table according to the UMI counts of each sample and percent of mitochondria rate to obtain the scaled data. PCA was constructed based on the scaled data with top 2000 high variable genes and top 10 principals were used for tSNE construction and UMAP construction. Utilizing graph-based cluster method, the unsupervised cell cluster result based the PCA top 10 principal was acquired and the marker genes were calculated by FindAllMarkers function with Wilcox rank sum test algorithm under following criteria: 1. lnFC > 0.25; 2. p-value < 0.05; 3. min.pct > 0.1. In order to identify the cell type detailed, the clusters of same cell type were selected for re-tSNE analysis, graph-based clustering and marker analysis. For pseudotime analysis, the Single-Cell Trajectories analysis utilizing Monocle2 (http://cole-trapnell-lab.github.io/monocle-release) using DDR-Tree and default parameter. Based on the pseudo-time analysis, branch expression analysis modeling (BEAM Analysis) was applied for branch fate determined gene analysis.
Statistics
Data are given as means ± SEM and analyzed with Prism version 6.0 (GraphPad Software). Data were analyzed by Student’s t-test between two groups. Survival curves were compared by log-rank (Mantel-Cox) test. Differences were considered significant when P < 0.05.
Study approval
The mouse care and experimental protocols conducted in this study were approved by the Huazhong University of Science and Technology Animal Care and Use Committee.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.